Somatic mosaicism and lineage tracing
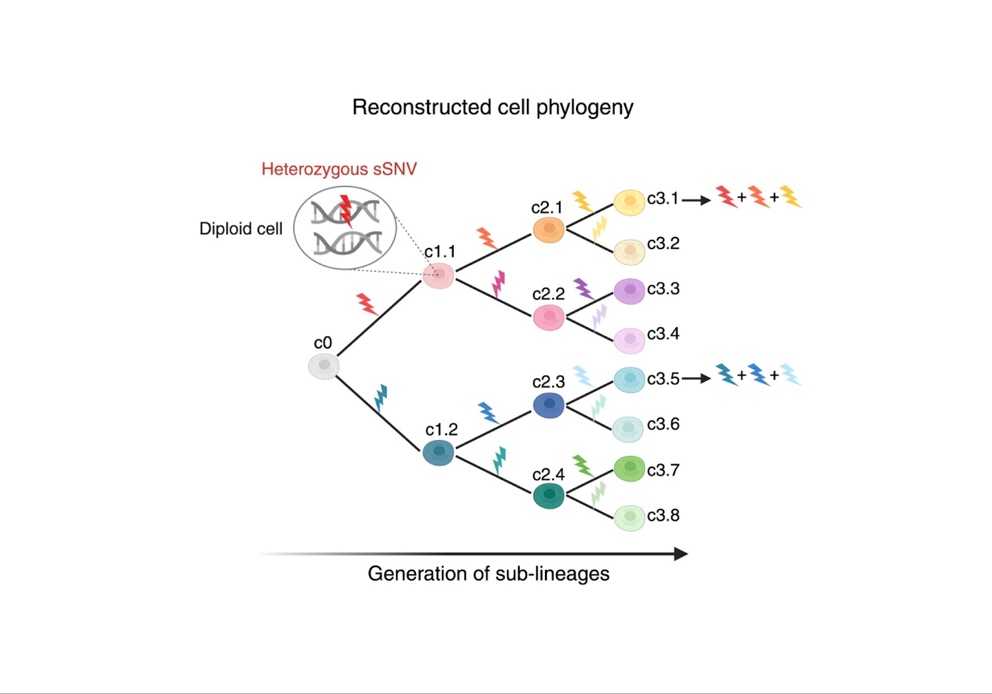 Development is a process of concerted cell generation and cell type specification. Progenitor cells proliferate to build different body parts, while tissues and cell types differentiate following the execution of precise gene expression programs. How cells are generated and distributed in our body is a key question in developmental biology. Successive cell divisions generate lineages (or clones) of related cells that can be resolved and visualized as a cell phylogeny in a process called lineage tracing. Lineage tracing is done through permanent, unique and cumulative tagging of progenitor cells, and allows to map and trace the cells during development. In animal and in vitro models, lineage tracing can be done by artificially introducing tags (or scars) in progenitor cells, for example through genome editing. In humans, such experiments cannot be done in vivo however, recent studies, have shown that post-zygotic somatic single nucleotide variants (sSNVs) that occur in progenitor cells at virtually every cell cycle, and that have no known pathological effect, can be used to perform retrospective lineage tracing starting directly from human tissue.
Development is a process of concerted cell generation and cell type specification. Progenitor cells proliferate to build different body parts, while tissues and cell types differentiate following the execution of precise gene expression programs. How cells are generated and distributed in our body is a key question in developmental biology. Successive cell divisions generate lineages (or clones) of related cells that can be resolved and visualized as a cell phylogeny in a process called lineage tracing. Lineage tracing is done through permanent, unique and cumulative tagging of progenitor cells, and allows to map and trace the cells during development. In animal and in vitro models, lineage tracing can be done by artificially introducing tags (or scars) in progenitor cells, for example through genome editing. In humans, such experiments cannot be done in vivo however, recent studies, have shown that post-zygotic somatic single nucleotide variants (sSNVs) that occur in progenitor cells at virtually every cell cycle, and that have no known pathological effect, can be used to perform retrospective lineage tracing starting directly from human tissue.
Cell lineages of human neurodevelopment
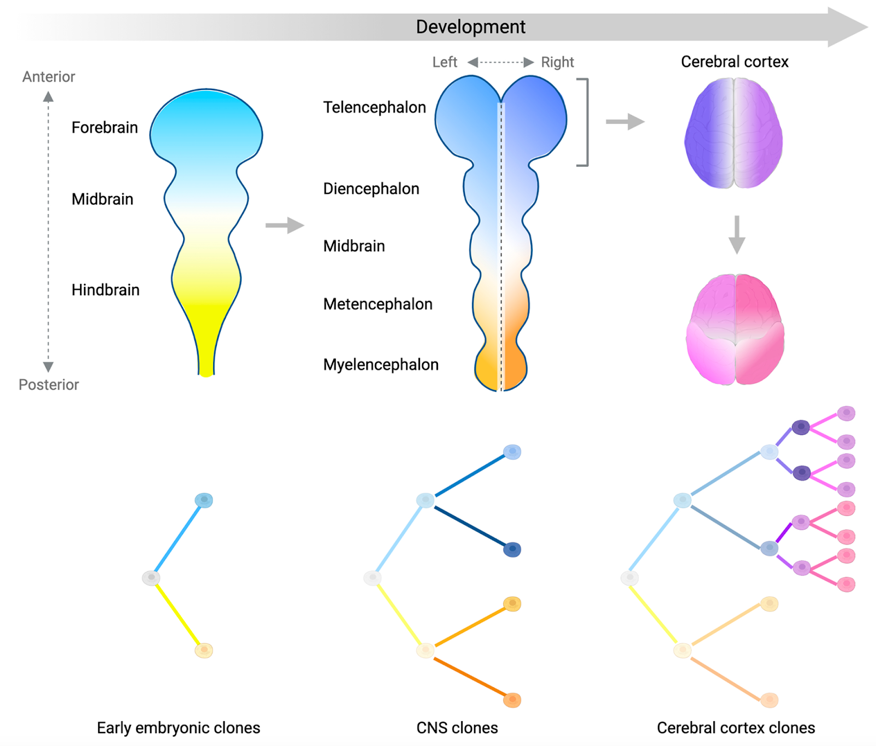 The human central nervous system (CNS) is a mosaic of >200 billion cells. CNS development starts with the formation of the neural tube after gastrulation, and ends with the most evolutionary expanded and complex of the brain structures, the cerebral cortex. How cells shape the CNS during development is a long-standing question in neurobiology. Human-specific structural features are only partially recapitulated in existing models, which stretches the need of conducting studies directly in humans. One of the objectives of our lab is to map and trace the developmental origin of the cells in the human CNS in order to understand its formation. For this, we use post-mortem human tissue, and apply cutting edge sequencing technologies including bulk and single-cell DNA sequencing and single-cell (multi)omics. Our lab also applies advanced computational tools and mathematical modeling to address these questions and interpret stochastic and deterministic processes acting on cell lineages and somatic evolution.
The human central nervous system (CNS) is a mosaic of >200 billion cells. CNS development starts with the formation of the neural tube after gastrulation, and ends with the most evolutionary expanded and complex of the brain structures, the cerebral cortex. How cells shape the CNS during development is a long-standing question in neurobiology. Human-specific structural features are only partially recapitulated in existing models, which stretches the need of conducting studies directly in humans. One of the objectives of our lab is to map and trace the developmental origin of the cells in the human CNS in order to understand its formation. For this, we use post-mortem human tissue, and apply cutting edge sequencing technologies including bulk and single-cell DNA sequencing and single-cell (multi)omics. Our lab also applies advanced computational tools and mathematical modeling to address these questions and interpret stochastic and deterministic processes acting on cell lineages and somatic evolution.
Pediatric brain disorders
Pediatric brain disorders include neurodevelopmental disorders such as cortical malformations, as well as low-grade and high-grade brain tumors. Cortical malformations form a wide spectrum of rare genetic, often non-syndromic developmental brain disorders associated with pharmaco-resistant epilepsy. While the genetic causes of some of these disorders are known, a significant fraction of cases (~45%) still remains genetically unsolved. Pediatric brain tumors, on the other hand, include Low-grade Epilepsy-Associated Tumors (LEATs) such as ganglioglioma (GG), Dysembryoplastic Neuroepithelial Tumors (DNTs) and pilocytic astrocytoma, and high-grade tumors such as Childhood Diffuse Intrinsic Pontine Glioma (DIPG), Atypical Teratoid Rhabdoid Tumor (ATRT), and embryonal brain tumors such as childhood medulloblastoma. Our lab works to 1) address the potential role of somatic variants in some of these disorders; 2) characterize the somatic genomic landscape of pediatric brain tumors, and 3) understand the origin of the disorders and their somatic development with a focus on cell lineages.








